First of all, read the user documentation to get accustomed with the main functionalities of Nemo. Then have a look at the code documentation presented here. Nemo is a project of moderate size, it contains less than 75 classes in its current state. This is enough to get lost if you don't know where to look first!
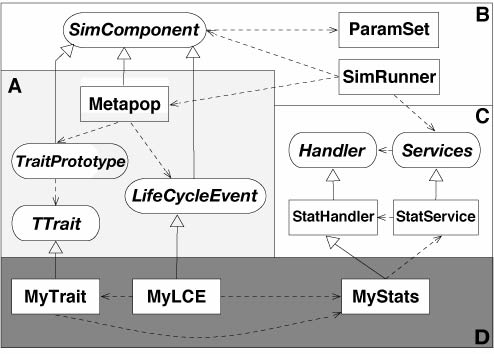
Simplified framework design |
Basic Classes:
The code is organized as follows: there are three main interface classes that a developer will use to derive his/her customized components. These are the LifeCycleEvent (LCE), TraitPrototype, and TTrait interfaces. Besides these three, the StatHandler and FileHandler interfaces will also be of importance when designing the output options of the components.
The TraitPrototype and LifeCycleEvent classes are derived from the SimComponent interface, a base class used to manage components' parameters and input/output functions. The TraitPrototype and TTrait classes also inherit from the StorableComponent interface, which declares methods to record components' data to a binary file.
Since the implementation of a GeneticMap, some of the traits also derive from the TTraitPotoWithMap interface, which handles recombination on a single genetic map shared by multiple trait classes.
Simulation components, and their parameters:
The main function of the SimComponent interface is to manage the set of parameters that the users will use to design and run a simulation. Each component of a simulation (LCE's and traits) thus declares a set of parameters stored in a ParamSet object within its SimComponent layer. In addition, each simulation component may declare an associated StatHandler to record summary statistics during the run of a simulation and associated output/input files whose behavior is defined by the appropriate FileHandler class. The SimComponent class provides a simple interface to make these stat and file handlers visible to the overall simulation stats and files managers, the StatServices and FileServices. These two Service classes maintain lists of the different Handler objects declared by all components of a simulation and notify them in time of action, that is, when statistics need to be recorded and stored (in a StatRecorder) or when trait data (e.g. genotypes) need to be written to a file.
The parameters of a simulation component must be defined during instanciation, that is, they must be added to the ParamSet member when the component constructor is called. The parameters are created and stored in the ParamSet object using the SimComponent interface ( SimComponent::add_parameter() ) and these instructions must be placed in the component's constructor (see Metapop::Metapop for an example).
Finally, the parameters are set from input user's values by a call to the SimComponent::setParameters abstract function, which must be defined when creating a new component (a trait or LCE). That mandatory function must hold the instructions how to interpret the parameter values from the input parameter file and initialize the component's internal variables (e.g., the number of loci and mutation rate of a trait). The setParameters function is called when the SimRunner initializes the simulation and reads the parameters from the input file(s) defined by the user.
Initialization sequence of the components:
The component manager holds a list of all components loaded at startup (the default components are loaded by SIMenv::loadDefaultComponents()). Your new components can be loaded after the SIMenv has loaded the default ones (see section five below). The sets of parameters of all components are then loaded into the parameter manager. At initialization, Nemo reads the parameters from the input file and asks the ParamManager to assign them to the right parameter set (the right SimComponent, a trait or LCE). Once this first initialization step is completed, the ComponentManager builds a list of the components that have been selected by the user's input parameters. A SimComponent is said to be selected when its ParamSet has all its mandatory parameters in the "set" state (i.e., its required Param members have been attributed a value of the right type). This ParamSet is then in the "isSet" state and the SimComponent that owns it can be loaded into the current list of simulation components that will be used to run the simulation. The LCEs and trait prototypes selected this way are then used to create the sequence of events in the life cycle and the Individual prototype. The prototype is then used to create all new individuals during the simulation by cloning it (see IndFactory). Finally, the stat and file services scan the selected components to load their handlers using the SimComponent interface.
The StorableComponent class declares the interface to save and load the component data to/from a binary file. It is thus used by the BinaryDataSaver and BinaryDataLoader classes.
Population framework:
The population framework gives access to traits within individuals that are themselves contained within patches aggregated within a metapopulation. The four main classes are thus Metapop, Patch, Individual, and TTrait. This hierarchy of classes is in fact a top-down chain of responsibility with the Metapop class being the top-class and the TTrait (derived) class(es), the bottom-class. The Metapop class provides the interface to access individuals and traits within the population. It also updates information about the population state such as number of individuals in each age class and patch, number of generations and replicates passed, etc. It further provides the interface to manipulate its own state, that is to move individuals between patches and/or age classes, or remove individuals/patches, and build a new population from scratch or by loading it from a binary file (see the Metapop class documentation).
The patches may also be directly accessed and manipulated through the Metapop interface. The Patch class provides the same kind of interface as the Metapop class do but limited to one sub-population only. You can thus get the number of individuals present within a particular patch in a given age class (see the Patch::size() functions), and add or remove individuals within a deme. Given the design pattern used in the population framework (i.e. the chain of responsibility pattern), patches are unaware of the state of the upper class and thus ignore the presence or absence of other patches within the population. They only know their own state and thus can only manipulate the elements they contain directly, that is the individuals. This is the reason why using the Patch interface one can not move an individual from a patch to another one. This is achieved within the Metapop interface. The same is true for the Individual class that maintains the trait container but ignores the state of other individuals or its own location within the population. Individuals are thus manipulated through the Patch interface to perform tasks such as mating and selection, or the Metapop interface to perform tasks that need spatial information like dispersal or spatial selection.
The Individual Factory: New individuals are created using the IndFactory base class of the Metapop class. This class maintains a garbage collector (called the recycling pool) for unused individuals and draw on it each time a new individual is needed. If the recycling pool is empty, a new individual is created by cloning the individual prototype (IndFactory::_protoIndividual) which contains the traits derived from the trait prototypes. The trait prototypes are themselves loaded to the IndFactory container (IndFactory::_protoTraits) at initialization of the metapopulation (using IndFactory::makePrototype()), that is, before a new simulation starts. The prototypes are used to create (instanciate) the traits loaded into the individual prototype. Those traits are then themselves cloned each time a new individual is created by cloning the individual prototype (when using IndFactory::getNewIndividual(), IndFactory::makeNewIndividual(), etc.). However, note that to create a new offspring from two parents, the IndFactory::makeOffsprg() member function has to be used in order to perform inheritance and mutation on the traits. The function IndFactory::makeNewIndividual() only allocates memory for the traits gene sequence when needed and sets the Individual inner properties. Inheritance and mutation can be performed by calling the Individual::create() member function directly with pointers to the mother and father as arguments.
The Traits: Traits are accessed within individuals using their index in the trait table of the Individual class (Individual::Traits). These indexes are stored in a hash table (std::map) within the IndFactory class and are thus accessible through the Metapop interface. The key used in the hash table is the trait's type (or name, accessed through TTrait::get_type()) and the index table is set when creating the individual prototype. The only way to access a trait within an individual (i.e. using Individual::getTrait()) is to know its index rather than its type. Therefore, the LCE interface provides the interface to set and store the index of a trait a particular LCE is using (see LifeCycleEvent::_LCELinkedTraitIndex and constructor). This is done at construction time of an LCE by passing the trait's type (a character string) as an argument to the LifeCycleEvent base class constructor. The trait index can also be set at runtime (see LCE_Breed_Selection::setParameters()) or more that one trait index may be used by one LCE (see LCE_Disperse_EvolDisp::setParameters()).
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
