Set patch-specifiec initial genotypes values. More...
#include <LCEquanti.h>
 Inheritance diagram for LCE_QuantiInit:
Inheritance diagram for LCE_QuantiInit: Collaboration diagram for LCE_QuantiInit:
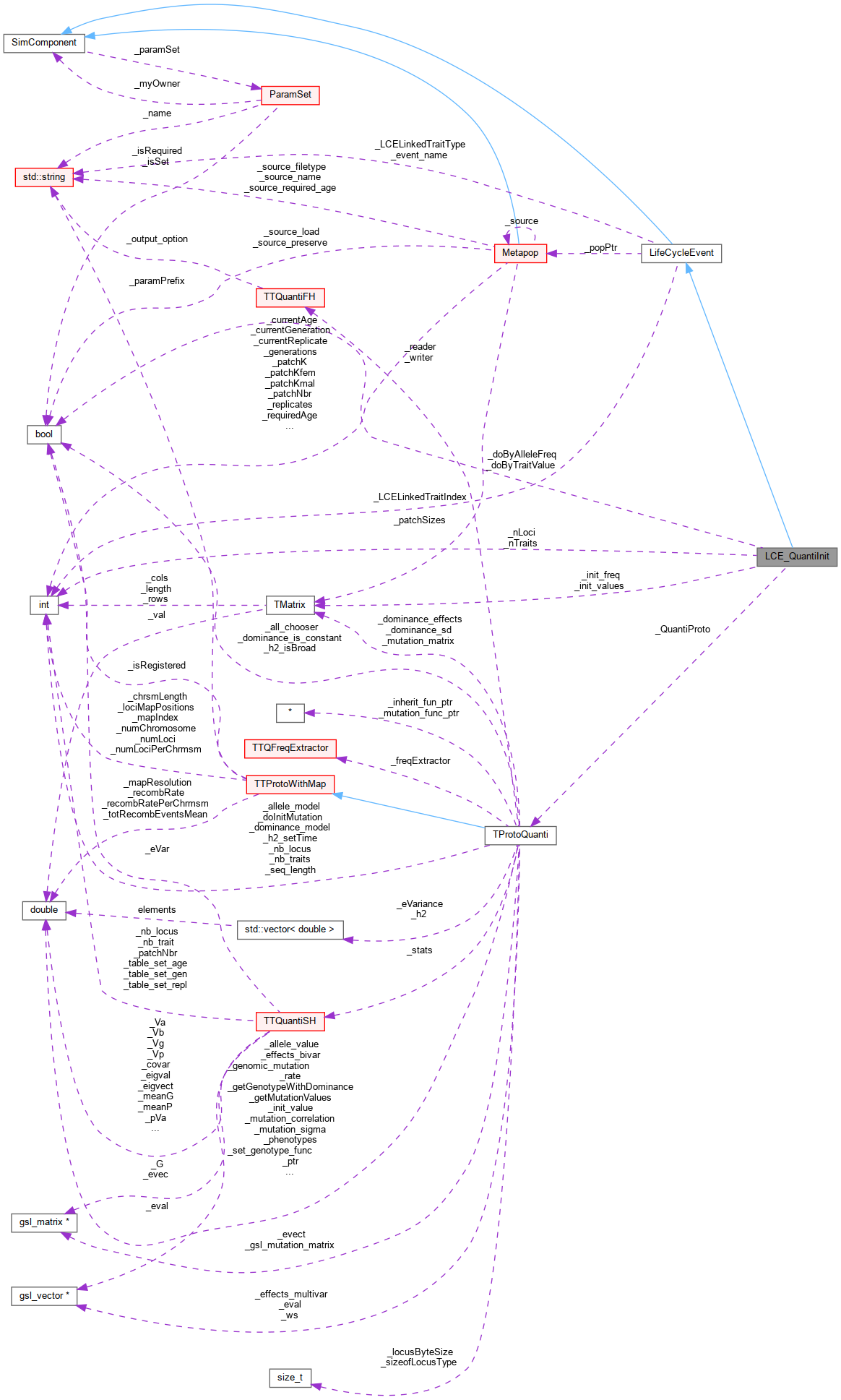
Collaboration diagram for LCE_QuantiInit:Public Member Functions | |
| LCE_QuantiInit () | |
| virtual | ~LCE_QuantiInit () |
| virtual void | execute () |
| void | init_trait_value (sex_t SEX, age_idx AGE, unsigned int size, unsigned int deme, double *values) |
| void | init_allele_freq (sex_t SEX, age_idx AGE, unsigned int size, unsigned int deme, double *values, const TMatrix &all_val) |
| virtual LifeCycleEvent * | clone () |
| virtual bool | setParameters () |
| virtual void | loadFileServices (FileServices *loader) |
| virtual void | loadStatServices (StatServices *loader) |
| virtual bool | resetParameterFromSource (std::string param, SimComponent *cmpt) |
| virtual age_t | removeAgeClass () |
| virtual age_t | addAgeClass () |
| virtual age_t | requiredAgeClass () |
 Public Member Functions inherited from LifeCycleEvent Public Member Functions inherited from LifeCycleEvent | |
| LifeCycleEvent (const char *name, const char *trait_link) | |
| Cstor. More... | |
| virtual | ~LifeCycleEvent () |
| virtual void | init (Metapop *popPtr) |
| Sets the pointer to the current Metapop and the trait link if applicable. More... | |
| virtual bool | attach_trait (string trait) |
| virtual void | set_paramset (std::string name, bool required, SimComponent *owner) |
| virtual void | set_event_name (std::string &name) |
| Set the name of the event (name of the ParamSet) and add the corresponding parameter to the set. More... | |
| virtual void | set_event_name (const char *name) |
| virtual string & | get_event_name () |
| Accessor to the LCE's name. More... | |
| virtual int | get_rank () |
| Accessor to the LCE rank in the life cycle. More... | |
| virtual void | set_pop_ptr (Metapop *popPtr) |
| Accessors for the population pointer. More... | |
| virtual Metapop * | get_pop_ptr () |
 Public Member Functions inherited from SimComponent Public Member Functions inherited from SimComponent | |
| SimComponent () | |
| virtual | ~SimComponent () |
| virtual void | loadUpdaters (UpdaterServices *loader) |
| Loads the parameters and component updater onto the updater manager. More... | |
| virtual void | set_paramset (ParamSet *paramset) |
| Sets the ParamSet member. More... | |
| virtual void | set_paramsetFromCopy (const ParamSet &PSet) |
| Reset the set of parameters from a another set. More... | |
| virtual ParamSet * | get_paramset () |
| ParamSet accessor. More... | |
| virtual void | add_parameter (Param *param) |
| Interface to add a parameter to the set. More... | |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd) |
| Interface to add a parameter to the set. More... | |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd, ParamUpdaterBase *updater) |
| Interface to add a parameter and its updater to the set. More... | |
| virtual Param * | get_parameter (std::string name) |
| Param getter. More... | |
| virtual double | get_parameter_value (std::string name) |
| Param value getter. More... | |
| virtual string | get_name () |
| Returnd the name of the ParamSet, i.e. More... | |
| virtual bool | has_parameter (std::string name) |
| Param getter. More... | |
Private Attributes | |
| TMatrix | _init_values |
| TMatrix | _init_freq |
| bool | _doByTraitValue |
| bool | _doByAlleleFreq |
| unsigned int | _nTraits |
| unsigned int | _nLoci |
| TProtoQuanti * | _QuantiProto |
Additional Inherited Members | |
 Protected Attributes inherited from LifeCycleEvent Protected Attributes inherited from LifeCycleEvent | |
| std::string | _event_name |
| The param name to be read in the init file. More... | |
| Metapop * | _popPtr |
| The ptr to the current Metapop. More... | |
| std::string | _LCELinkedTraitType |
| The name of the linked trait. More... | |
| int | _LCELinkedTraitIndex |
| The index in the individual's trait table of the linked trait. More... | |
 Protected Attributes inherited from SimComponent Protected Attributes inherited from SimComponent | |
| ParamSet * | _paramSet |
| The parameters container. More... | |
Detailed Description
Set patch-specifiec initial genotypes values.
Constructor & Destructor Documentation
◆ LCE_QuantiInit()
| LCE_QuantiInit::LCE_QuantiInit | ( | ) |
References SimComponent::add_parameter(), and MAT.
Referenced by clone().
◆ ~LCE_QuantiInit()
Member Function Documentation
◆ addAgeClass()
|
inlinevirtual |
Implements LifeCycleEvent.
◆ clone()
|
inlinevirtual |
◆ execute()
|
virtual |
Implements LifeCycleEvent.
References _doByAlleleFreq, _doByTraitValue, _init_freq, _init_values, _nLoci, _nTraits, LifeCycleEvent::_popPtr, _QuantiProto, ADLTx, ADULTS, FEM, Metapop::getCurrentGeneration(), Metapop::getPatchNbr(), TMatrix::getRowView(), IndFactory::getTraitPrototype(), init_allele_freq(), init_trait_value(), MAL, message(), OFFSPRG, OFFSx, TProtoQuanti::set_init_values(), and Metapop::size().
◆ init_allele_freq()
| void LCE_QuantiInit::init_allele_freq | ( | sex_t | SEX, |
| age_idx | AGE, | ||
| unsigned int | size, | ||
| unsigned int | deme, | ||
| double * | values, | ||
| const TMatrix & | all_val | ||
| ) |
References LifeCycleEvent::_LCELinkedTraitIndex, _nLoci, LifeCycleEvent::_popPtr, Metapop::get(), Individual::getTrait(), RAND::SampleSeqWithReciprocal(), TTQuanti_diallelic::set_allele_bit(), Individual::setTraitValue(), and RAND::Uniform().
Referenced by execute().
◆ init_trait_value()
| void LCE_QuantiInit::init_trait_value | ( | sex_t | SEX, |
| age_idx | AGE, | ||
| unsigned int | size, | ||
| unsigned int | deme, | ||
| double * | values | ||
| ) |
References LifeCycleEvent::_LCELinkedTraitIndex, LifeCycleEvent::_popPtr, Metapop::get(), Individual::getTrait(), TTrait::init_sequence(), and TTrait::set_value().
Referenced by execute().
◆ loadFileServices()
|
inlinevirtual |
Implements SimComponent.
◆ loadStatServices()
|
inlinevirtual |
Implements SimComponent.
◆ removeAgeClass()
|
inlinevirtual |
Implements LifeCycleEvent.
◆ requiredAgeClass()
|
inlinevirtual |
Implements LifeCycleEvent.
◆ resetParameterFromSource()
|
inlinevirtual |
Implements SimComponent.
◆ setParameters()
|
virtual |
Implements SimComponent.
References _doByAlleleFreq, _doByTraitValue, _init_freq, _init_values, _nLoci, _nTraits, SimComponent::_paramSet, LifeCycleEvent::_popPtr, _QuantiProto, error(), fatal(), TProtoQuanti::get_allele_model(), TProtoQuanti::get_num_locus(), SimComponent::get_parameter(), ParamSet::getMatrix(), Metapop::getPatchNbr(), IndFactory::getTraitPrototype(), ParamSet::isSet(), setSpatialMatrix(), and warning().
Member Data Documentation
◆ _doByAlleleFreq
|
private |
Referenced by execute(), and setParameters().
◆ _doByTraitValue
|
private |
Referenced by execute(), and setParameters().
◆ _init_freq
|
private |
Referenced by execute(), and setParameters().
◆ _init_values
|
private |
Referenced by execute(), and setParameters().
◆ _nLoci
|
private |
Referenced by execute(), init_allele_freq(), and setParameters().
◆ _nTraits
|
private |
Referenced by execute(), and setParameters().
◆ _QuantiProto
|
private |
Referenced by execute(), and setParameters().
The documentation for this class was generated from the following files:
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
