Microsatellites genome.
More...
#include <ttneutralgenes.h>
◆ TTNeutralGenes() [1/2]
| TTNeutralGenes::TTNeutralGenes |
( |
| ) |
|
|
inline |
TProtoNeutralGenes * _myProto
Definition: ttneutralgenes.h:58
void(TTNeutralGenes::* _mutate_func_ptr)(void)
Definition: ttneutralgenes.h:60
const trait_t _type
Definition: ttneutralgenes.h:66
unsigned char ** _sequence
Definition: ttneutralgenes.h:64
void(TProtoNeutralGenes::* _inherit_func_ptr)(sex_t, unsigned char *, const unsigned char **)
Definition: ttneutralgenes.h:61
#define NTRL
Definition: types.h:70
Referenced by clone().
◆ TTNeutralGenes() [2/2]
◆ ~TTNeutralGenes()
| TTNeutralGenes::~TTNeutralGenes |
( |
| ) |
|
|
virtual |
◆ clone()
◆ get_allele_value()
| double TTNeutralGenes::get_allele_value |
( |
int |
loc, |
|
|
int |
all |
|
) |
| const |
|
virtual |
◆ get_sequence()
| virtual void** TTNeutralGenes::get_sequence |
( |
| ) |
const |
|
inlinevirtual |
◆ get_type()
| virtual trait_t TTNeutralGenes::get_type |
( |
| ) |
const |
|
inlinevirtual |
◆ getValue()
| virtual void* TTNeutralGenes::getValue |
( |
| ) |
const |
|
inlinevirtual |
◆ inherit()
| void TTNeutralGenes::inherit |
( |
const TTrait * |
mother, |
|
|
const TTrait * |
father |
|
) |
| |
|
virtual |
◆ init()
| void TTNeutralGenes::init |
( |
| ) |
|
|
virtual |
◆ init_sequence()
| void TTNeutralGenes::init_sequence |
( |
| ) |
|
|
virtual |
◆ mutate()
| virtual void TTNeutralGenes::mutate |
( |
| ) |
|
|
inlinevirtual |
◆ mutate_2all()
| void TTNeutralGenes::mutate_2all |
( |
| ) |
|
|
inline |
◆ mutate_KAM()
| void TTNeutralGenes::mutate_KAM |
( |
| ) |
|
◆ mutate_NULL()
| void TTNeutralGenes::mutate_NULL |
( |
| ) |
|
|
inline |
◆ mutate_SSM()
| void TTNeutralGenes::mutate_SSM |
( |
| ) |
|
◆ operator!=()
| bool TTNeutralGenes::operator!= |
( |
const TTrait & |
T | ) |
|
|
virtual |
◆ operator=()
◆ operator==()
| bool TTNeutralGenes::operator== |
( |
const TTrait & |
T | ) |
|
|
virtual |
◆ reset()
| void TTNeutralGenes::reset |
( |
| ) |
|
|
virtual |
◆ retrieve_data()
◆ set_allele()
| void TTNeutralGenes::set_allele |
( |
unsigned int |
loc, |
|
|
unsigned int |
al, |
|
|
unsigned char |
val |
|
) |
| |
|
inline |
◆ set_allele_value()
| void TTNeutralGenes::set_allele_value |
( |
unsigned int |
locus, |
|
|
unsigned int |
allele, |
|
|
double |
value |
|
) |
| |
|
virtual |
Implements TTrait.
550 assert(locus < _myProto->get_locus_num() && allele < 2);
551 _sequence[allele][locus] = (
unsigned char)value;
References _sequence.
◆ set_inherit_func_ptr()
| void TTNeutralGenes::set_inherit_func_ptr |
( |
void(TProtoNeutralGenes::*)(sex_t, unsigned char *, const unsigned char **) |
theFunc | ) |
|
|
inline |
◆ set_mut_func_ptr()
| void TTNeutralGenes::set_mut_func_ptr |
( |
void(TTNeutralGenes::*)(void) |
theFunc | ) |
|
|
inline |
◆ set_proto()
◆ set_sequence()
| void TTNeutralGenes::set_sequence |
( |
void ** |
seq | ) |
|
|
virtual |
◆ set_trait()
| virtual void* TTNeutralGenes::set_trait |
( |
void * |
value | ) |
|
|
inlinevirtual |
◆ set_value()
| virtual void TTNeutralGenes::set_value |
( |
| ) |
|
|
inlinevirtual |
◆ show_up()
| void TTNeutralGenes::show_up |
( |
| ) |
|
|
virtual |
◆ store_data()
◆ _inherit_func_ptr
| void(TProtoNeutralGenes::* TTNeutralGenes::_inherit_func_ptr) (sex_t, unsigned char *, const unsigned char **) |
|
private |
◆ _mutate_func_ptr
| void(TTNeutralGenes::* TTNeutralGenes::_mutate_func_ptr) (void) |
|
private |
◆ _myProto
Referenced by get_allele_value(), inherit(), init(), init_sequence(), mutate_2all(), mutate_KAM(), mutate_SSM(), operator=(), operator==(), reset(), retrieve_data(), set_proto(), set_sequence(), show_up(), store_data(), and ~TTNeutralGenes().
◆ _sequence
| unsigned char** TTNeutralGenes::_sequence |
|
private |
Referenced by get_allele_value(), get_sequence(), inherit(), init(), init_sequence(), mutate_2all(), mutate_KAM(), mutate_SSM(), operator=(), reset(), retrieve_data(), set_allele(), set_allele_value(), set_sequence(), show_up(), store_data(), and ~TTNeutralGenes().
◆ _type
| const trait_t TTNeutralGenes::_type |
|
private |
The documentation for this class was generated from the following files:
 Inheritance diagram for TTNeutralGenes:
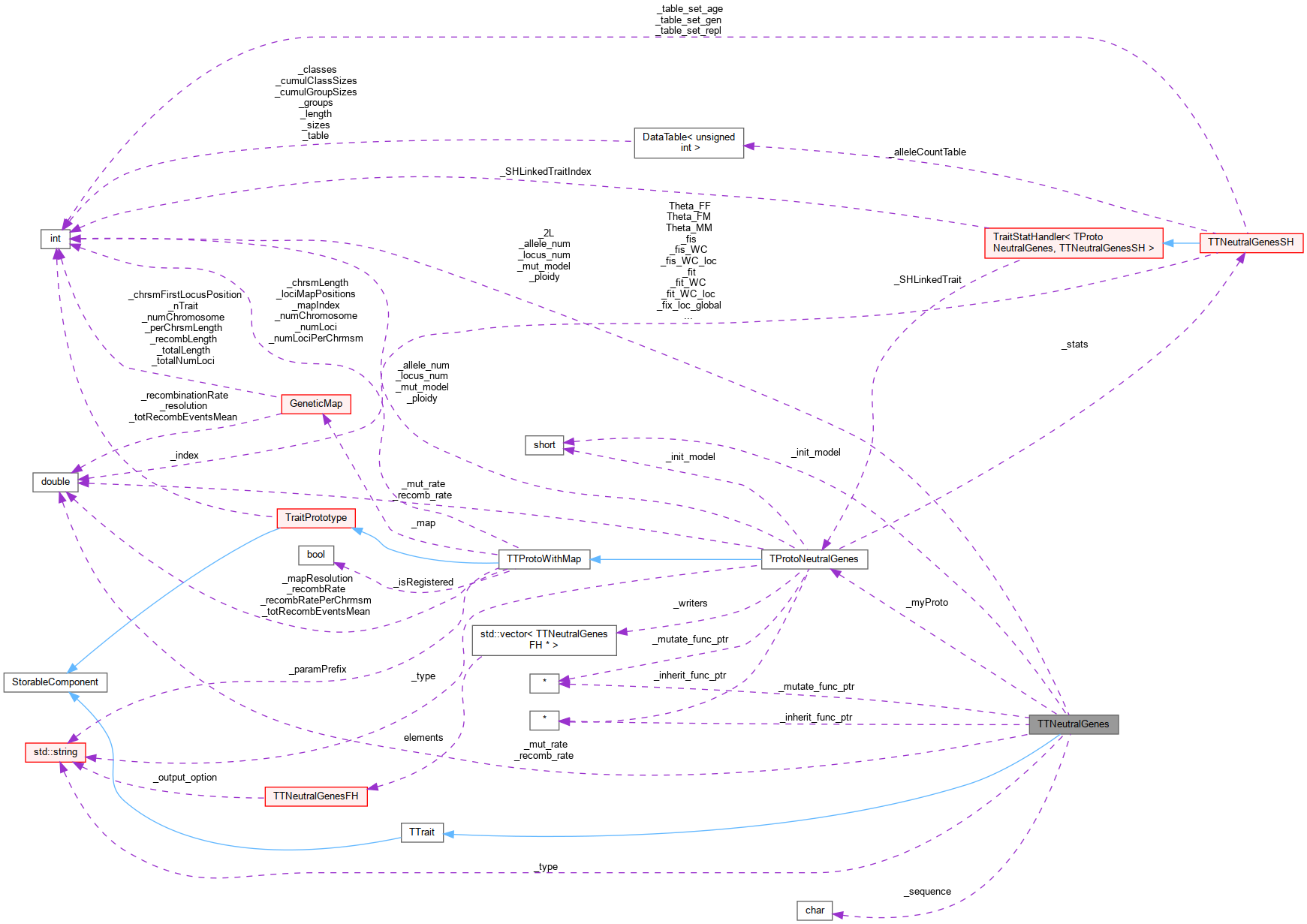
Inheritance diagram for TTNeutralGenes: Collaboration diagram for TTNeutralGenes:
Collaboration diagram for TTNeutralGenes: Public Member Functions inherited from TTrait
Public Member Functions inherited from TTrait Public Member Functions inherited from StorableComponent
Public Member Functions inherited from StorableComponent
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
