Prototype class for the TTNeutralGenes trait class. More...
#include <ttneutralgenes.h>
 Inheritance diagram for TProtoNeutralGenes:
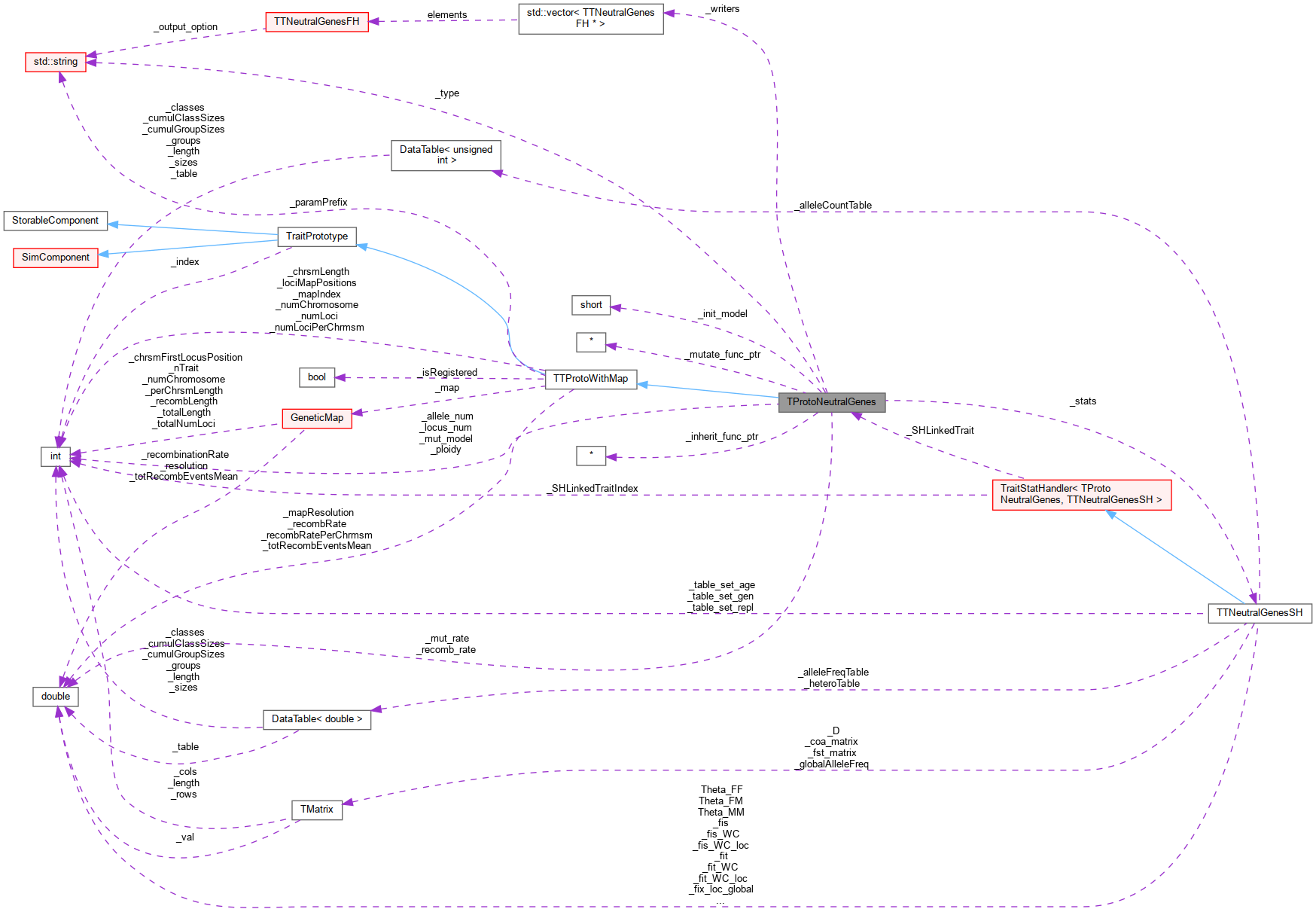
Inheritance diagram for TProtoNeutralGenes: Collaboration diagram for TProtoNeutralGenes:
Collaboration diagram for TProtoNeutralGenes:Public Member Functions | |
| TProtoNeutralGenes () | |
| TProtoNeutralGenes (const TProtoNeutralGenes &T) | |
| virtual | ~TProtoNeutralGenes () |
| unsigned int | get_ploidy () |
| unsigned int | get_locus_num () |
| unsigned int | get_allele_num () |
| double | get_mut_rate () |
| unsigned int | get_genome_size () |
| unsigned int | get_num_mutations () |
| unsigned int | get_init_model () |
| TTNeutralGenesSH * | get_stater () |
| void | inherit_low (sex_t SEX, unsigned char *seq, const unsigned char **parent) |
| void | inherit_free (sex_t SEX, unsigned char *seq, const unsigned char **parent) |
| virtual void | init () |
| virtual void | reset () |
| virtual TTNeutralGenes * | hatch () |
| virtual TProtoNeutralGenes * | clone () |
| virtual trait_t | get_type () const |
| virtual bool | setParameters () |
| virtual void | loadFileServices (FileServices *loader) |
| virtual void | loadStatServices (StatServices *loader) |
| virtual bool | resetParameterFromSource (std::string param, SimComponent *cmpt) |
| virtual void | store_data (BinaryStorageBuffer *saver) |
| virtual bool | retrieve_data (BinaryStorageBuffer *reader) |
 Public Member Functions inherited from TTProtoWithMap Public Member Functions inherited from TTProtoWithMap | |
| TTProtoWithMap () | |
| TTProtoWithMap (const TTProtoWithMap &TP) | |
| virtual | ~TTProtoWithMap () |
| void | setMapIndex (unsigned int idx) |
| unsigned int | getMapIndex () |
| bool | setGeneticMapParameters (string prefix) |
| void | addGeneticMapParameters (string prefix) |
| bool | setRecombinationMapRandom () |
| bool | setRecombinationMapNonRandom (vector< vector< double > > *lociPositions) |
| bool | setRecombinationMapFixed () |
| bool | setNumLociPerChromosome (string param_name) |
| void | reset_recombination_pointers () |
| void | registerGeneticMap () |
| void | unregisterFromGeneticMap () |
| bool | areGeneticMapParamSet (string prefix) |
| bool | isRecombinationFree (string prefix) |
| void | recordRandomMap () |
 Public Member Functions inherited from TraitPrototype Public Member Functions inherited from TraitPrototype | |
| virtual void | set_index (int idx) |
| Sets the traits index. More... | |
| virtual int | get_index () |
| Index getter. More... | |
 Public Member Functions inherited from StorableComponent Public Member Functions inherited from StorableComponent | |
| virtual | ~StorableComponent () |
 Public Member Functions inherited from SimComponent Public Member Functions inherited from SimComponent | |
| SimComponent () | |
| virtual | ~SimComponent () |
| virtual void | loadUpdaters (UpdaterServices *loader) |
| Loads the parameters and component updater onto the updater manager. More... | |
| virtual void | set_paramset (ParamSet *paramset) |
| Sets the ParamSet member. More... | |
| virtual void | set_paramset (std::string name, bool required, SimComponent *owner) |
| Sets a new ParamSet and name it. More... | |
| virtual void | set_paramsetFromCopy (const ParamSet &PSet) |
| Reset the set of parameters from a another set. More... | |
| virtual ParamSet * | get_paramset () |
| ParamSet accessor. More... | |
| virtual void | add_parameter (Param *param) |
| Interface to add a parameter to the set. More... | |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd) |
| Interface to add a parameter to the set. More... | |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd, ParamUpdaterBase *updater) |
| Interface to add a parameter and its updater to the set. More... | |
| virtual Param * | get_parameter (std::string name) |
| Param getter. More... | |
| virtual double | get_parameter_value (std::string name) |
| Param value getter. More... | |
| virtual string | get_name () |
| Returnd the name of the ParamSet, i.e. More... | |
| virtual bool | has_parameter (std::string name) |
| Param getter. More... | |
Private Attributes | |
| unsigned int | _allele_num |
| unsigned int | _locus_num |
| unsigned int | _ploidy |
| double | _mut_rate |
| int | _mut_model |
| unsigned int | _2L |
| unsigned int | _init_model |
| double | _recomb_rate |
| void(TTNeutralGenes::* | _mutate_func_ptr )(void) |
| void(TProtoNeutralGenes::* | _inherit_func_ptr )(sex_t, unsigned char *, const unsigned char **) |
| vector< TTNeutralGenesFH * > | _writers |
| TTNOhtaStats * | _ohtaStats |
| TTNeutralGenesSH * | _stats |
| const trait_t | _type |
Additional Inherited Members | |
 Static Public Member Functions inherited from TTProtoWithMap Static Public Member Functions inherited from TTProtoWithMap | |
| static void | recombine (unsigned long indID) |
 Static Public Attributes inherited from TTProtoWithMap Static Public Attributes inherited from TTProtoWithMap | |
| static GeneticMap | _map |
 Protected Attributes inherited from TTProtoWithMap Protected Attributes inherited from TTProtoWithMap | |
| unsigned int | _mapIndex |
| double | _totRecombEventsMean |
| double | _recombRate |
| double | _mapResolution |
| unsigned int | _numChromosome |
| unsigned int | _numLoci |
| double * | _recombRatePerChrmsm |
| unsigned int * | _numLociPerChrmsm |
| unsigned int * | _chrsmLength |
| unsigned int * | _lociMapPositions |
 Protected Attributes inherited from TraitPrototype Protected Attributes inherited from TraitPrototype | |
| int | _index |
| The trait index in the Individual traits table. More... | |
 Protected Attributes inherited from SimComponent Protected Attributes inherited from SimComponent | |
| ParamSet * | _paramSet |
| The parameters container. More... | |
Detailed Description
Prototype class for the TTNeutralGenes trait class.
Constructor & Destructor Documentation
◆ TProtoNeutralGenes() [1/2]
| TProtoNeutralGenes::TProtoNeutralGenes | ( | ) |
References SimComponent::add_parameter(), TTProtoWithMap::addGeneticMapParameters(), BOOL, DBL, INT, SimComponent::set_paramset(), and STR.
Referenced by clone().
◆ TProtoNeutralGenes() [2/2]
| TProtoNeutralGenes::TProtoNeutralGenes | ( | const TProtoNeutralGenes & | T | ) |
References SimComponent::_paramSet.
◆ ~TProtoNeutralGenes()
|
virtual |
References _ohtaStats, _stats, and _writers.
Member Function Documentation
◆ clone()
|
inlinevirtual |
◆ get_allele_num()
|
inline |
References _allele_num.
Referenced by TTNeutralGenesFH::FHread(), TTNOhtaStats::FHwrite(), TTNeutralGenesSH::init(), TTNeutralGenes::init_sequence(), TTNeutralGenes::mutate_KAM(), TTNeutralGenes::mutate_SSM(), TTNeutralGenes::operator==(), TTNeutralGenesSH::setAlleleTables(), TTNeutralGenesSH::setFreqRecorders(), TTNeutralGenesSH::setFst_li(), TTNeutralGenesSH::setFstat(), TTNeutralGenesSH::setFstat2(), TTNeutralGenesSH::setFstatWeirCockerham(), TTNeutralGenesSH::setFstatWeirCockerham_MS(), TTNeutralGenesSH::setFstMatrix(), TTNeutralGenesSH::setHeteroTable(), TTNeutralGenesSH::setHeterozygosity(), TTNeutralGenesSH::setHs(), TTNeutralGenesSH::setHs2(), TTNeutralGenesSH::setHt(), TTNeutralGenesSH::setHt2(), TTNeutralGenesSH::setLociDivCounter(), TTNeutralGenesSH::setNeiGeneticDistance(), LCE_NtrlInit::setParameters(), TTNeutralGenes::show_up(), TTNeutralGenesFH::write_Fst_i(), TTNeutralGenesFH::write_FSTAT(), TTNeutralGenesFH::write_GENEPOP(), TTNeutralGenesFH::write_PLINK(), and TTNeutralGenesFH::write_varcompWC().
◆ get_genome_size()
◆ get_init_model()
|
inline |
◆ get_locus_num()
|
inline |
References _locus_num.
Referenced by TTNeutralGenesFH::FHread(), TTNOhtaStats::FHwrite(), TTNeutralGenes::get_allele_value(), TTNeutralGenesSH::getDxyPerPatch(), TTNeutralGenes::init(), TTNeutralGenesSH::init(), TTNeutralGenes::init_sequence(), TTNeutralGenes::mutate_2all(), TTNeutralGenes::mutate_KAM(), TTNeutralGenes::mutate_SSM(), TTNeutralGenes::operator=(), TTNeutralGenes::operator==(), TTNeutralGenesFH::print_PLINK_PED(), TTNeutralGenes::retrieve_data(), TTNeutralGenes::set_sequence(), TTNeutralGenesSH::setAlleleTables(), TTNeutralGenesSH::setFreqRecorders(), TTNeutralGenesSH::setFst_li(), TTNeutralGenesSH::setFstat(), TTNeutralGenesSH::setFstat2(), TTNeutralGenesSH::setFstatWeirCockerham(), TTNeutralGenesSH::setFstatWeirCockerham_MS(), TTNeutralGenesSH::setFstMatrix(), TTNeutralGenesSH::setHeteroTable(), TTNeutralGenesSH::setHeterozygosity(), TTNeutralGenesSH::setHo(), TTNeutralGenesSH::setHo2(), TTNeutralGenesSH::setHs(), TTNeutralGenesSH::setHs2(), TTNeutralGenesSH::setHt(), TTNeutralGenesSH::setHt2(), TTNeutralGenesSH::setLociDivCounter(), TTNeutralGenesSH::setNeiGeneticDistance(), LCE_NtrlInit::setParameters(), TTNeutralGenes::show_up(), TTNeutralGenes::store_data(), TTNeutralGenesFH::write_Fst_i(), TTNeutralGenesFH::write_FSTAT(), TTNeutralGenesFH::write_GENEPOP(), TTNeutralGenesFH::write_patch_FSTAT(), TTNeutralGenesFH::write_patch_GENEPOP(), TTNeutralGenesFH::write_patch_TAB(), TTNeutralGenesFH::write_PLINK(), TTNeutralGenesFH::write_TAB(), and TTNeutralGenesFH::write_varcompWC().
◆ get_mut_rate()
◆ get_num_mutations()
|
inline |
References _2L, _mut_rate, and RAND::Binomial().
Referenced by TTNeutralGenes::mutate_2all(), TTNeutralGenes::mutate_KAM(), and TTNeutralGenes::mutate_SSM().
◆ get_ploidy()
|
inline |
References _ploidy.
Referenced by TTNeutralGenes::get_allele_value(), TTNeutralGenes::init_sequence(), TTNeutralGenes::operator==(), TTNeutralGenesFH::print_PLINK_PED(), TTNeutralGenes::reset(), TTNeutralGenes::retrieve_data(), TTNeutralGenes::store_data(), TTNeutralGenesFH::write_FSTAT(), TTNeutralGenesFH::write_GENEPOP(), TTNeutralGenesFH::write_patch_FSTAT(), TTNeutralGenesFH::write_patch_GENEPOP(), TTNeutralGenesFH::write_patch_TAB(), TTNeutralGenesFH::write_PLINK(), TTNeutralGenesFH::write_TAB(), and TTNeutralGenes::~TTNeutralGenes().
◆ get_stater()
|
inline |
References _stats.
Referenced by TTNeutralGenesFH::write_Fst_i(), and TTNeutralGenesFH::write_varcompWC().
◆ get_type()
|
inlinevirtual |
◆ hatch()
|
virtual |
Implements TraitPrototype.
References _inherit_func_ptr, _mutate_func_ptr, TTNeutralGenes::set_inherit_func_ptr(), TTNeutralGenes::set_mut_func_ptr(), and TTNeutralGenes::set_proto().
◆ inherit_free()
|
inline |
◆ inherit_low()
|
inline |
References TTProtoWithMap::_map, TTProtoWithMap::_mapIndex, and GeneticMap::reduceJunctions().
Referenced by setParameters().
◆ init()
◆ loadFileServices()
|
virtual |
Implements SimComponent.
References _allele_num, _ohtaStats, _writers, FileServices::attach(), FileServices::attach_reader(), fatal(), SimComponent::get_parameter(), Param::getArg(), Param::getMatrix(), Param::getValue(), Param::isMatrix(), Param::isSet(), SIMenv::MainSim, TraitFileHandler< TP >::set(), FileHandler::set_extension(), FileHandler::set_isInputHandler(), TraitFileHandler< TP >::set_multi(), TTNeutralGenesFH::set_write_fct(), TTNeutralGenesFH::setOutputOption(), warning(), TTNeutralGenesFH::write_Fst_i(), TTNeutralGenesFH::write_FSTAT(), TTNeutralGenesFH::write_GENEPOP(), TTNeutralGenesFH::write_PLINK(), TTNeutralGenesFH::write_TAB(), and TTNeutralGenesFH::write_varcompWC().
◆ loadStatServices()
|
virtual |
Implements SimComponent.
References _stats, and StatServices::attach().
◆ reset()
|
inlinevirtual |
◆ resetParameterFromSource()
|
virtual |
Implements SimComponent.
References error(), SimComponent::get_parameter(), and Param::isSet().
◆ retrieve_data()
|
virtual |
Implements StorableComponent.
References _locus_num, BinaryStorageBuffer::read(), and warning().
◆ setParameters()
|
virtual |
Implements SimComponent.
References _2L, _allele_num, _inherit_func_ptr, _init_model, _locus_num, _mut_model, _mut_rate, _mutate_func_ptr, _ploidy, TTProtoWithMap::_recombRate, error(), SimComponent::get_parameter(), SimComponent::get_parameter_value(), inherit_free(), inherit_low(), TTProtoWithMap::isRecombinationFree(), Param::isSet(), TTNeutralGenes::mutate_2all(), TTNeutralGenes::mutate_KAM(), TTNeutralGenes::mutate_NULL(), TTNeutralGenes::mutate_SSM(), and TTProtoWithMap::setGeneticMapParameters().
◆ store_data()
|
inlinevirtual |
Implements StorableComponent.
References _locus_num, and BinaryStorageBuffer::store().
Member Data Documentation
◆ _2L
|
private |
Referenced by get_genome_size(), get_num_mutations(), and setParameters().
◆ _allele_num
|
private |
Referenced by get_allele_num(), loadFileServices(), and setParameters().
◆ _inherit_func_ptr
|
private |
Referenced by hatch(), and setParameters().
◆ _init_model
|
private |
Referenced by get_init_model(), and setParameters().
◆ _locus_num
|
private |
Referenced by get_locus_num(), inherit_free(), retrieve_data(), setParameters(), and store_data().
◆ _mut_model
|
private |
Referenced by setParameters().
◆ _mut_rate
|
private |
Referenced by get_mut_rate(), get_num_mutations(), and setParameters().
◆ _mutate_func_ptr
|
private |
Referenced by hatch(), and setParameters().
◆ _ohtaStats
|
private |
Referenced by loadFileServices(), and ~TProtoNeutralGenes().
◆ _ploidy
|
private |
Referenced by get_ploidy(), and setParameters().
◆ _recomb_rate
|
private |
◆ _stats
|
private |
Referenced by get_stater(), loadStatServices(), and ~TProtoNeutralGenes().
◆ _type
|
private |
Referenced by get_type().
◆ _writers
|
private |
Referenced by loadFileServices(), and ~TProtoNeutralGenes().
The documentation for this class was generated from the following files:
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
