|
| | TTProtoWithMap () |
| |
| | TTProtoWithMap (const TTProtoWithMap &TP) |
| |
| virtual | ~TTProtoWithMap () |
| |
| void | setMapIndex (unsigned int idx) |
| |
| unsigned int | getMapIndex () |
| |
| bool | setGeneticMapParameters (string prefix) |
| |
| void | addGeneticMapParameters (string prefix) |
| |
| bool | setRecombinationMapRandom () |
| |
| bool | setRecombinationMapNonRandom (vector< vector< double > > *lociPositions) |
| |
| bool | setRecombinationMapFixed () |
| |
| bool | setNumLociPerChromosome (string param_name) |
| |
| void | reset_recombination_pointers () |
| |
| void | registerGeneticMap () |
| |
| void | unregisterFromGeneticMap () |
| |
| bool | areGeneticMapParamSet (string prefix) |
| |
| bool | isRecombinationFree (string prefix) |
| |
| void | recordRandomMap () |
| |
| virtual void | reset () |
| |
| virtual TTrait * | hatch ()=0 |
| | Creates the trait of which it is the prototype, called by IndFactory::makePrototype(). More...
|
| |
| virtual TraitPrototype * | clone ()=0 |
| | Returns a copy of itself. More...
|
| |
| virtual trait_t | get_type () const =0 |
| | Type accessor. More...
|
| |
| virtual void | set_index (int idx) |
| | Sets the traits index. More...
|
| |
| virtual int | get_index () |
| | Index getter. More...
|
| |
| virtual void | store_data (BinaryStorageBuffer *saver)=0 |
| | Interface to store the component data (e.g. gene values) into a binary buffer. More...
|
| |
| virtual bool | retrieve_data (BinaryStorageBuffer *reader)=0 |
| | Interface to retrieve the same data from the binary buffer. More...
|
| |
| virtual | ~StorableComponent () |
| |
| | SimComponent () |
| |
| virtual | ~SimComponent () |
| |
| virtual void | loadFileServices (FileServices *loader)=0 |
| | Loads the component's FileHandler onto the FileServices. More...
|
| |
| virtual void | loadStatServices (StatServices *loader)=0 |
| | Loads the component's StatHandler onto the StatServices. More...
|
| |
| virtual void | loadUpdaters (UpdaterServices *loader) |
| | Loads the parameters and component updater onto the updater manager. More...
|
| |
| virtual bool | setParameters ()=0 |
| | Default interface needed to initialize the component's variables from its input parameters value. More...
|
| |
| virtual void | set_paramset (ParamSet *paramset) |
| | Sets the ParamSet member. More...
|
| |
| virtual void | set_paramset (std::string name, bool required, SimComponent *owner) |
| | Sets a new ParamSet and name it. More...
|
| |
| virtual void | set_paramsetFromCopy (const ParamSet &PSet) |
| | Reset the set of parameters from a another set. More...
|
| |
| virtual ParamSet * | get_paramset () |
| | ParamSet accessor. More...
|
| |
| virtual void | add_parameter (Param *param) |
| | Interface to add a parameter to the set. More...
|
| |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd) |
| | Interface to add a parameter to the set. More...
|
| |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd, ParamUpdaterBase *updater) |
| | Interface to add a parameter and its updater to the set. More...
|
| |
| virtual Param * | get_parameter (std::string name) |
| | Param getter. More...
|
| |
| virtual double | get_parameter_value (std::string name) |
| | Param value getter. More...
|
| |
| virtual string | get_name () |
| | Returnd the name of the ParamSet, i.e. More...
|
| |
| virtual bool | has_parameter (std::string name) |
| | Param getter. More...
|
| |
| virtual bool | resetParameterFromSource (std::string param, SimComponent *cmpt)=0 |
| |
| bool TTProtoWithMap::setGeneticMapParameters |
( |
string |
prefix | ) |
|
128 bool map_set =
false;
137 param_name = prefix +
"_genetic_map_resolution";
147 param_name = prefix +
"_recombination_rate";
168 return error(
"\"%s_recombination_rate\" must be one-dimensional, with chromosome-specific recombination rates as elements.\n", prefix.c_str());
195 param_name = prefix +
"_genetic_map";
199 return error(
"A genetic map is already specified for trait \"%s\" while parameter \"%s\" is set.\n",prefix.c_str(), param_name.c_str());
202 vector< vector<double> > tmp_varmatx;
208 param_name = prefix +
"_chromosome_num_locus";
211 warning(
"\"%s_chromosome_num_locus\" is not used with \"%s_genetic_map\", ignoring it.\n", prefix.c_str(), prefix.c_str());
218 unsigned int length = 0;
226 return error(
"Number of loci in \"%s_genetic_map\" (%i) different from number of loci (%i) for trait \"%s\"\n",
227 prefix.c_str(), length,
_numLoci, prefix.c_str());
237 param_name = prefix +
"_random_genetic_map";
241 return error(
"A genetic map is already specified for trait \"%s\" while parameter \"%s\" is set.\n",param_name.c_str());
247 error(
"\"%s_random_genetic_map\" must have one row, with chromosome lengths as elements.\n",
get_type().c_str());
void getVariableMatrix(vector< vector< double > > *mat)
Definition: param.cc:460
void getMatrix(TMatrix *mat)
Sets the matrix from the argument string if the parameter is set and of matrix type.
Definition: param.cc:378
bool isSet()
Definition: param.h:140
A class to handle matrix in params, coerces matrix into a vector of same total size.
Definition: tmatrix.h:50
unsigned int getNbRows() const
Gives the number of rows.
Definition: tmatrix.h:212
unsigned int getNbCols() const
Gives the number of columns.
Definition: tmatrix.h:215
double get(unsigned int i, unsigned int j) const
Accessor to element at row i and column j.
Definition: tmatrix.h:193
bool setNumLociPerChromosome(string param_name)
Definition: ttrait_with_map.cc:297
bool setRecombinationMapFixed()
Definition: ttrait_with_map.cc:388
void registerGeneticMap()
Definition: ttrait_with_map.cc:575
bool setRecombinationMapNonRandom(vector< vector< double > > *lociPositions)
Definition: ttrait_with_map.cc:364
bool setRecombinationMapRandom()
Definition: ttrait_with_map.cc:438
int error(const char *str,...)
Definition: output.cc:77
void warning(const char *str,...)
Definition: output.cc:58
References _chrsmLength, _mapResolution, _numChromosome, _numLoci, _numLociPerChrmsm, _paramPrefix, _recombRate, _recombRatePerChrmsm, error(), TMatrix::get(), SimComponent::get_parameter(), SimComponent::get_parameter_value(), TraitPrototype::get_type(), Param::getMatrix(), TMatrix::getNbCols(), TMatrix::getNbRows(), Param::getVariableMatrix(), Param::isSet(), registerGeneticMap(), reset_recombination_pointers(), setNumLociPerChromosome(), setRecombinationMapFixed(), setRecombinationMapNonRandom(), setRecombinationMapRandom(), and warning().
Referenced by TProtoQuanti::setGeneticMapParams(), TProtoBDMI::setParameters(), TProtoDeletMutations_bitstring::setParameters(), and TProtoNeutralGenes::setParameters().
| bool TTProtoWithMap::setRecombinationMapRandom |
( |
| ) |
|
!there might be duplicates, we don't deal with those
452 vector< unsigned int > tmp_map;
453 unsigned int **chrm_map;
468 vector<unsigned int>::iterator vec_first = tmp_map.begin(), vec_last = tmp_map.end();
470 std::sort(vec_first, vec_last);
475 chrm_map[i][j] = tmp_map[j];
495 delete [] chrm_map[i];
518 mapParam->
setArg(map.str());
static double Uniform()
Generates a random number from [0.0, 1.0[ uniformly distributed.
Definition: Uniform.h:124
References _chrsmLength, _lociMapPositions, _mapResolution, _numChromosome, _numLoci, _numLociPerChrmsm, _paramPrefix, tstring::dble2str(), SimComponent::get_parameter(), Param::setArg(), Param::setIsSet(), and RAND::Uniform().
Referenced by setGeneticMapParameters().
 Inheritance diagram for TTProtoWithMap:
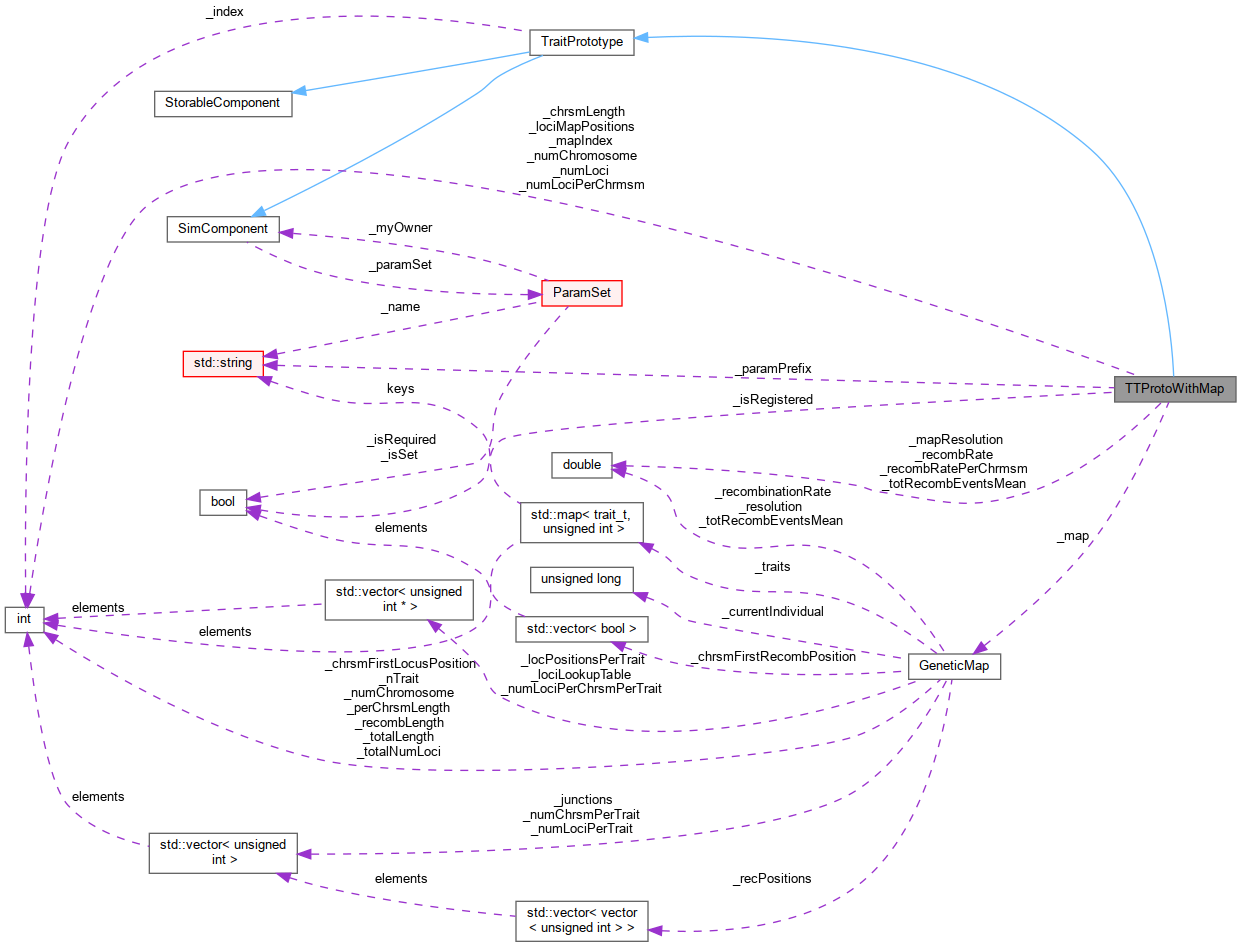
Inheritance diagram for TTProtoWithMap: Collaboration diagram for TTProtoWithMap:
Collaboration diagram for TTProtoWithMap: Public Member Functions inherited from TraitPrototype
Public Member Functions inherited from TraitPrototype Public Member Functions inherited from StorableComponent
Public Member Functions inherited from StorableComponent Public Member Functions inherited from SimComponent
Public Member Functions inherited from SimComponent Protected Attributes inherited from TraitPrototype
Protected Attributes inherited from TraitPrototype Protected Attributes inherited from SimComponent
Protected Attributes inherited from SimComponent
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
