|
| | TProtoBDMI () |
| |
| | TProtoBDMI (const TProtoBDMI &TP) |
| |
| virtual | ~TProtoBDMI () |
| |
| int | get_num_locus () |
| |
| double | get_mut_rate () |
| |
| bool | isHaploid () |
| |
| void | set_init_freq (double *val, unsigned int size) |
| |
| double | get_init_freq (unsigned int i) |
| |
| bool | isInitSet () |
| |
| double | getGenoFitnessHaplo (unsigned int row, unsigned int pos) |
| |
| double | getGenoFitnessDiplo (unsigned int row, unsigned int posA, unsigned int posB) |
| |
| double | getGenoFitnessDiplo (unsigned int row, unsigned int pos) |
| |
| void | setGenoFitnessValue (unsigned int row, unsigned int geno, double value) |
| |
| void | showGenoTable (unsigned int nrows) |
| |
| void | gamete_recombination (sex_t SEX, bitstring &seq, const bitstring *parent) |
| |
| void | inherit_free (sex_t SEX, bitstring &seq, const bitstring *parent) |
| |
| void | inherit_low (sex_t SEX, bitstring &seq, const bitstring *parent) |
| |
|
|
| virtual void | init () |
| |
| virtual TTrait * | hatch () |
| |
| virtual TraitPrototype * | clone () |
| |
| virtual trait_t | get_type () const |
| |
| virtual void | store_data (BinaryStorageBuffer *saver) |
| |
| virtual bool | retrieve_data (BinaryStorageBuffer *reader) |
| |
| virtual bool | setParameters () |
| |
| virtual void | loadFileServices (FileServices *loader) |
| |
| virtual void | loadStatServices (StatServices *loader) |
| |
| virtual bool | resetParameterFromSource (std::string param, SimComponent *cmpt) |
| |
| | TTProtoWithMap () |
| |
| | TTProtoWithMap (const TTProtoWithMap &TP) |
| |
| virtual | ~TTProtoWithMap () |
| |
| void | setMapIndex (unsigned int idx) |
| |
| unsigned int | getMapIndex () |
| |
| bool | setGeneticMapParameters (string prefix) |
| |
| void | addGeneticMapParameters (string prefix) |
| |
| bool | setRecombinationMapRandom () |
| |
| bool | setRecombinationMapNonRandom (vector< vector< double > > *lociPositions) |
| |
| bool | setRecombinationMapFixed () |
| |
| bool | setNumLociPerChromosome (string param_name) |
| |
| void | reset_recombination_pointers () |
| |
| void | registerGeneticMap () |
| |
| void | unregisterFromGeneticMap () |
| |
| bool | areGeneticMapParamSet (string prefix) |
| |
| bool | isRecombinationFree (string prefix) |
| |
| void | recordRandomMap () |
| |
| virtual void | reset () |
| |
| virtual void | set_index (int idx) |
| | Sets the traits index. More...
|
| |
| virtual int | get_index () |
| | Index getter. More...
|
| |
| virtual | ~StorableComponent () |
| |
| | SimComponent () |
| |
| virtual | ~SimComponent () |
| |
| virtual void | loadUpdaters (UpdaterServices *loader) |
| | Loads the parameters and component updater onto the updater manager. More...
|
| |
| virtual void | set_paramset (ParamSet *paramset) |
| | Sets the ParamSet member. More...
|
| |
| virtual void | set_paramset (std::string name, bool required, SimComponent *owner) |
| | Sets a new ParamSet and name it. More...
|
| |
| virtual void | set_paramsetFromCopy (const ParamSet &PSet) |
| | Reset the set of parameters from a another set. More...
|
| |
| virtual ParamSet * | get_paramset () |
| | ParamSet accessor. More...
|
| |
| virtual void | add_parameter (Param *param) |
| | Interface to add a parameter to the set. More...
|
| |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd) |
| | Interface to add a parameter to the set. More...
|
| |
| virtual void | add_parameter (std::string Name, param_t Type, bool isRequired, bool isBounded, double low_bnd, double up_bnd, ParamUpdaterBase *updater) |
| | Interface to add a parameter and its updater to the set. More...
|
| |
| virtual Param * | get_parameter (std::string name) |
| | Param getter. More...
|
| |
| virtual double | get_parameter_value (std::string name) |
| | Param value getter. More...
|
| |
| virtual string | get_name () |
| | Returnd the name of the ParamSet, i.e. More...
|
| |
| virtual bool | has_parameter (std::string name) |
| | Param getter. More...
|
| |
|
| static unsigned int | _diploGenotTableCoding3x3 [3][3] = {{0,1,2},{3,4,5},{6,7,8}} |
| |
| static unsigned int | _diploGenotTableCoding4x4 [4][4] = {{0,1,2,3},{4,5,6,7},{8,9,10,11},{12,13,14,15}} |
| |
| bool TProtoBDMI::setParameters |
( |
| ) |
|
|
virtual |
Implements SimComponent.
91 return error(
"dmi trait: the number of loci must be an even number; incompatible loci go by pair!\n");
118 return error(
"dmi trait: the genotype fitness table must have 4 elements per locus pair in the haploid case.");
122 return error(
"dmi trait: the genotype fitness table must have 9 or 16 elements per locus pair in the diploid case.");
126 return error(
"dmi trait: the number of rows of the genotype fitness table exceeds the number of locus pairs.\n");
145 unsigned int index[16] = {0,1,1,2,3,4,4,5,3,4,4,5,6,7,7,8};
154 for(
unsigned int i = 0; i < oldmat.nrows(); ++i)
155 for(
unsigned int j = 0; j < 16; ++j)
163 message(
"::TProtoBDMI::\nFitness table:\n");
virtual double get_parameter_value(std::string name)
Param value getter.
Definition: simcomponent.h:143
void reset(unsigned int rows, unsigned int cols)
Re-allocate the existing matrix with assigned rows and cols dimensions and all elements to 0.
Definition: tmatrix.h:161
unsigned int ncols() const
Definition: tmatrix.h:216
void copy_recycle(const TMatrix &mat)
Copy elements of 'mat', recycling elements of 'mat' if its size is smaller than current matrix.
Definition: tmatrix.h:90
unsigned int nrows() const
Definition: tmatrix.h:213
void inherit_free(sex_t SEX, bitstring &seq, const bitstring *parent)
Definition: ttbdmi.cc:288
void inherit_low(sex_t SEX, bitstring &seq, const bitstring *parent)
Definition: ttbdmi.cc:297
void showGenoTable(unsigned int nrows)
Definition: ttbdmi.cc:261
double _recombRate
Definition: ttrait_with_map.h:195
bool isRecombinationFree(string prefix)
Definition: ttrait_with_map.cc:98
bool setGeneticMapParameters(string prefix)
Definition: ttrait_with_map.cc:125
void mutate_diplo()
Definition: ttbdmi.cc:580
double viability_diplo()
Definition: ttbdmi.cc:617
double viability_haplo()
Definition: ttbdmi.cc:603
void inherit_haplo(const TTrait *mother, const TTrait *father)
Definition: ttbdmi.cc:546
void mutate_haplo()
Definition: ttbdmi.cc:564
void inherit_diplo(const TTrait *mother, const TTrait *father)
Definition: ttbdmi.cc:535
int error(const char *str,...)
Definition: output.cc:77
void message(const char *message,...)
Definition: output.cc:40
References _gamete_recomb_func_ptr, _genomic_mut_rate, _genoTable, _inherit_func_ptr, _isHaploid, _isInitSet, _mut_rate, _mutation_func_ptr, _npair, _num_locus, TTProtoWithMap::_recombRate, _viability_func_ptr, TMatrix::copy_recycle(), error(), TMatrix::get(), SimComponent::get_parameter(), SimComponent::get_parameter_value(), Param::getMatrix(), TT_BDMI::inherit_diplo(), inherit_free(), TT_BDMI::inherit_haplo(), inherit_low(), TTProtoWithMap::isRecombinationFree(), message(), TT_BDMI::mutate_diplo(), TT_BDMI::mutate_haplo(), TMatrix::ncols(), TMatrix::nrows(), TMatrix::reset(), TMatrix::set(), TTProtoWithMap::setGeneticMapParameters(), showGenoTable(), TT_BDMI::viability_diplo(), and TT_BDMI::viability_haplo().
Referenced by init().
 Inheritance diagram for TProtoBDMI:
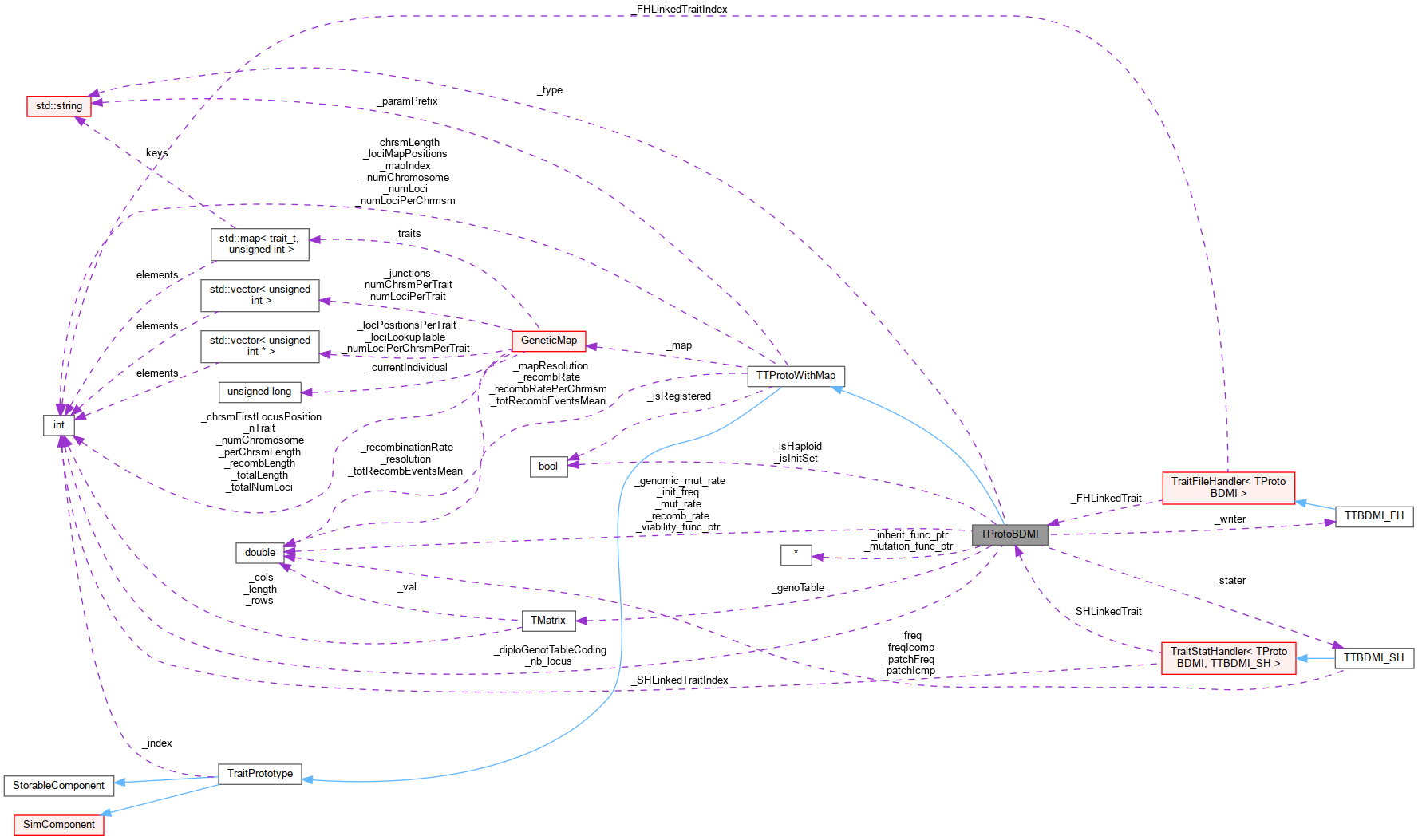
Inheritance diagram for TProtoBDMI: Collaboration diagram for TProtoBDMI:
Collaboration diagram for TProtoBDMI: Public Member Functions inherited from TTProtoWithMap
Public Member Functions inherited from TTProtoWithMap Public Member Functions inherited from TraitPrototype
Public Member Functions inherited from TraitPrototype Public Member Functions inherited from StorableComponent
Public Member Functions inherited from StorableComponent Public Member Functions inherited from SimComponent
Public Member Functions inherited from SimComponent Static Public Member Functions inherited from TTProtoWithMap
Static Public Member Functions inherited from TTProtoWithMap Static Public Attributes inherited from TTProtoWithMap
Static Public Attributes inherited from TTProtoWithMap Protected Attributes inherited from TTProtoWithMap
Protected Attributes inherited from TTProtoWithMap Protected Attributes inherited from TraitPrototype
Protected Attributes inherited from TraitPrototype Protected Attributes inherited from SimComponent
Protected Attributes inherited from SimComponent
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
