#include <ttbdmi.h>
 Inheritance diagram for TT_BDMI:
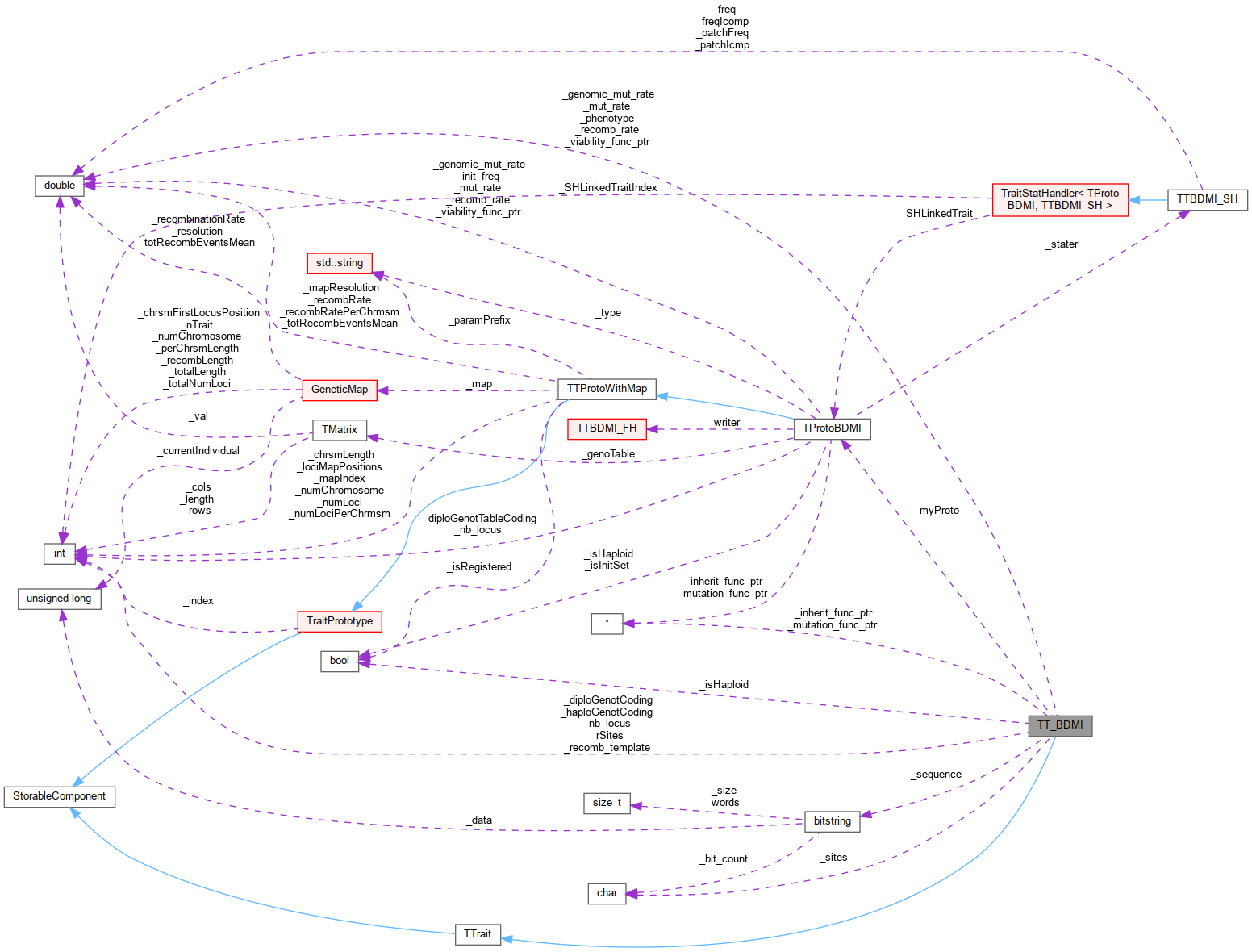
Inheritance diagram for TT_BDMI: Collaboration diagram for TT_BDMI:
Collaboration diagram for TT_BDMI:Public Member Functions | |
| TT_BDMI () | |
| TT_BDMI (const TT_BDMI &T) | |
| virtual | ~TT_BDMI () |
| void | set_sequence (bitstring **seq) |
| void | inherit_haplo (const TTrait *mother, const TTrait *father) |
| void | inherit_diplo (const TTrait *mother, const TTrait *father) |
| void | mutate_haplo () |
| void | mutate_diplo () |
| double | viability_haplo () |
| double | viability_diplo () |
| unsigned int | get_num_mut_haplo (unsigned int loc) |
| unsigned int | get_num_mut_diplo (unsigned int loc) |
| const bitstring & | get_bit_sequence (bool chromosome) const |
| const bitstring * | get_genome_sequence () const |
Setters: | |
| void | set_proto (TProtoBDMI *proto) |
Implementations | |
| virtual void | init () |
| virtual void | init_sequence () |
| virtual void | reset () |
| virtual void | inherit (const TTrait *mother, const TTrait *father) |
| virtual void | mutate () |
| virtual void * | set_trait (void *value) |
| virtual void | set_sequence (void **seq) |
| virtual void | set_value () |
| virtual void * | getValue () const |
| virtual trait_t | get_type () const |
| virtual void ** | get_sequence () const |
| virtual double | get_allele_value (int loc, int all) const |
| virtual void | set_allele_value (unsigned int locus, unsigned int allele, double value) |
| virtual void | show_up () |
| virtual TT_BDMI * | clone () |
| virtual TT_BDMI & | operator= (const TTrait &) |
| virtual bool | operator== (const TTrait &) |
| virtual bool | operator!= (const TTrait &) |
| virtual void | store_data (BinaryStorageBuffer *saver) |
| virtual bool | retrieve_data (BinaryStorageBuffer *reader) |
 Public Member Functions inherited from TTrait Public Member Functions inherited from TTrait | |
| virtual | ~TTrait () |
 Public Member Functions inherited from StorableComponent Public Member Functions inherited from StorableComponent | |
| virtual | ~StorableComponent () |
Private Attributes | |
| TProtoBDMI * | _myProto |
| bitstring | _sequence [2] |
| double | _phenotype |
Static Private Attributes | |
| static unsigned int | _haploGenotCoding [2][2] = {{0,1},{2,3}} |
| static unsigned int | _diploGenotCoding [2][2] = {{0,1},{2,3}} |
Constructor & Destructor Documentation
◆ TT_BDMI() [1/2]
|
inline |
Referenced by clone().
◆ TT_BDMI() [2/2]
|
inline |
◆ ~TT_BDMI()
Member Function Documentation
◆ clone()
◆ get_allele_value()
|
virtual |
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::getGenoFitnessDiplo(), and TProtoBDMI::getGenoFitnessHaplo().
◆ get_bit_sequence()
|
inline |
◆ get_genome_sequence()
|
inline |
References _sequence.
Referenced by inherit_diplo(), TTBDMI_FH::write_diplo(), and TTBDMI_FH::write_haplo().
◆ get_num_mut_diplo()
|
inline |
References _sequence.
Referenced by TTBDMI_SH::countAllele_diplo().
◆ get_num_mut_haplo()
|
inline |
◆ get_sequence()
|
inlinevirtual |
◆ get_type()
|
inlinevirtual |
◆ getValue()
|
inlinevirtual |
Implements TTrait.
References _phenotype.
Referenced by show_up(), TTBDMI_FH::write_diplo(), and TTBDMI_FH::write_haplo().
◆ inherit()
Implements TTrait.
References TProtoBDMI::_inherit_func_ptr, and _myProto.
◆ inherit_diplo()
References _myProto, _sequence, FEM, TProtoBDMI::gamete_recombination(), get_genome_sequence(), and MAL.
Referenced by TProtoBDMI::setParameters().
◆ inherit_haplo()
References _myProto, _sequence, TProtoBDMI::gamete_recombination(), and get_bit_sequence().
Referenced by TProtoBDMI::setParameters().
◆ init()
|
virtual |
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::_num_locus, _sequence, and bitstring::reset().
Referenced by operator=(), and set_sequence().
◆ init_sequence()
|
virtual |
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::_num_locus, _sequence, TProtoBDMI::get_init_freq(), TProtoBDMI::isInitSet(), bitstring::reset(), bitstring::set(), and RAND::Uniform().
Referenced by LCE_Init_BDMI::init_value().
◆ mutate()
|
inlinevirtual |
◆ mutate_diplo()
|
inline |
References TProtoBDMI::_genomic_mut_rate, _myProto, TProtoBDMI::_num_locus, _sequence, bitstring::flip(), RAND::Poisson(), RAND::RandBool(), and RAND::Uniform().
Referenced by TProtoBDMI::setParameters().
◆ mutate_haplo()
|
inline |
References TProtoBDMI::_genomic_mut_rate, _myProto, TProtoBDMI::_num_locus, _sequence, bitstring::flip(), RAND::Poisson(), and RAND::Uniform().
Referenced by TProtoBDMI::setParameters().
◆ operator!=()
◆ operator=()
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::_num_locus, _sequence, bitstring::copy(), init(), reset(), and set_value().
◆ operator==()
|
virtual |
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::_num_locus, TProtoBDMI::get_type(), and TTrait::get_type().
◆ reset()
|
virtual |
Implements TTrait.
References _sequence, and bitstring::reset().
Referenced by operator=(), and set_sequence().
◆ retrieve_data()
|
virtual |
Implements StorableComponent.
References TProtoBDMI::_isHaploid, _myProto, _sequence, bitstring::nb_words(), BinaryStorageBuffer::read(), and bitstring::set_data().
◆ set_allele_value()
|
virtual |
Implements TTrait.
References _myProto, and TProtoBDMI::setGenoFitnessValue().
◆ set_proto()
|
inline |
◆ set_sequence() [1/2]
| void TT_BDMI::set_sequence | ( | bitstring ** | seq | ) |
References TProtoBDMI::_isHaploid, _myProto, _sequence, bitstring::copy(), init(), and reset().
◆ set_sequence() [2/2]
|
inlinevirtual |
Implements TTrait.
◆ set_trait()
|
inlinevirtual |
Implements TTrait.
◆ set_value()
|
inlinevirtual |
Implements TTrait.
References _myProto, _phenotype, and TProtoBDMI::_viability_func_ptr.
Referenced by LCE_Init_BDMI::init_value(), operator=(), and show_up().
◆ show_up()
|
virtual |
Implements TTrait.
References TProtoBDMI::_isHaploid, _myProto, TProtoBDMI::_npair, TProtoBDMI::_num_locus, _sequence, getValue(), set_value(), and TProtoBDMI::showGenoTable().
◆ store_data()
|
virtual |
Implements StorableComponent.
References TProtoBDMI::_isHaploid, _myProto, _sequence, bitstring::getword_atIdx(), bitstring::nb_words(), and BinaryStorageBuffer::store().
◆ viability_diplo()
|
inline |
References _diploGenotCoding, _myProto, TProtoBDMI::_npair, TProtoBDMI::_num_locus, _sequence, and TProtoBDMI::getGenoFitnessDiplo().
Referenced by TProtoBDMI::setParameters().
◆ viability_haplo()
|
inline |
References _haploGenotCoding, _myProto, TProtoBDMI::_npair, TProtoBDMI::_num_locus, _sequence, and TProtoBDMI::getGenoFitnessHaplo().
Referenced by TProtoBDMI::setParameters().
Member Data Documentation
◆ _diploGenotCoding
|
staticprivate |
Referenced by viability_diplo().
◆ _haploGenotCoding
|
staticprivate |
Referenced by viability_haplo().
◆ _myProto
|
private |
Referenced by get_allele_value(), get_type(), inherit(), inherit_diplo(), inherit_haplo(), init(), init_sequence(), mutate(), mutate_diplo(), mutate_haplo(), operator=(), operator==(), retrieve_data(), set_allele_value(), set_proto(), set_sequence(), set_value(), show_up(), store_data(), viability_diplo(), and viability_haplo().
◆ _phenotype
|
private |
Referenced by getValue(), and set_value().
◆ _sequence
|
private |
Referenced by get_bit_sequence(), get_genome_sequence(), get_num_mut_diplo(), get_num_mut_haplo(), inherit_diplo(), inherit_haplo(), init(), init_sequence(), mutate_diplo(), mutate_haplo(), operator=(), reset(), retrieve_data(), set_sequence(), show_up(), store_data(), viability_diplo(), and viability_haplo().
The documentation for this class was generated from the following files:
 1.9.1 -- Nemo is hosted on
1.9.1 -- Nemo is hosted on 
